As the push for exposome research grows, more examples are needed of studies that have successfully captured cumulative exposures and related biological responses while considering the baseline genetic background. In a recent paper published in Current Environmental Health Reports, Chirag Patel, publisher of the first environmental-wide association study (EWAS), describes the challenges of exposome characterization at a population level and offers immediate solutions along with a larger vision for tackling the analytical complexities of gene variant-by-environment exposure interactions.
Genome x exposome analysis is necessary for understanding key exposures that matter to health at the population level. Although initiatives in personalized medicine will primarily offer a window into exposures that matter to small subsets of individuals, exposome research offers an opportunity to more broadly change the paradigm around health and disease.
In his article “Analytical Complexity in Detection of Gene Variant-by-Environment Exposure Interactions in High-Throughput Genomic and Exposomic Research”, Dr. Patel provides a background for EWAS with a brief review of the methods used for its closest sibling, the GWAS (genome-wide association study). What quickly becomes clear is the sample size needed to properly power an analysis for environmental exposures (particularly low prevalence exposures) and minimize risk of false positives and false negatives is not something readily accessible (even a well-characterized cohort of 10,000 would have limited power for this level of analysis).
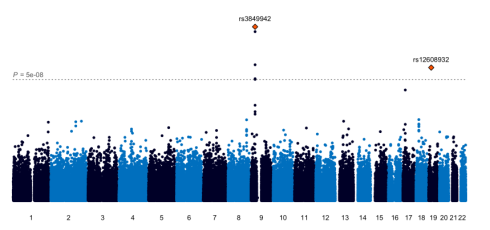 Using data from his original EWAS, the paper provides examples of how individual GWAS and EWAS analyses can be completed to first identify the most significant/biologically meaningful signatures that should then be included in a later GxE analysis. Multiple open access tools have been developed through his research group to aid in this process. He additionally suggests data-driven incorporation of biological and/or epidemiological findings through utilizing tools like the Comparative Toxicogenomics Database and Toxic Exposome Database to inform the discovery of potential GxE interactions. Although the analytical challenges of genome x exposome interactions are only beginning, the article offers viable solutions to continue to move exposome research forward. Read the full journal article here.
Using data from his original EWAS, the paper provides examples of how individual GWAS and EWAS analyses can be completed to first identify the most significant/biologically meaningful signatures that should then be included in a later GxE analysis. Multiple open access tools have been developed through his research group to aid in this process. He additionally suggests data-driven incorporation of biological and/or epidemiological findings through utilizing tools like the Comparative Toxicogenomics Database and Toxic Exposome Database to inform the discovery of potential GxE interactions. Although the analytical challenges of genome x exposome interactions are only beginning, the article offers viable solutions to continue to move exposome research forward. Read the full journal article here.
